Bayes Classifier example
BayesClassifier.RmdPrepare a simulated dataset for a Bayes Classifier
In the first step, we simulate a dataset with the following specifications:
- a total number of 10 classes, 4 of which are known and 6 are unknown, respectively
- n_labeled = 200 labeled samples, evenly distributed over the 4 known classes
- n_unlabeled = 5000 unlabeled samples, evenly distributed over the 10 classes
- a dimension of n_feats = 2 features (to facilitate representation)
Each class \(i\) is represented by a bivariate Gaussian distribution with mean vector \(\mu = (\mu_1^{(i)}, \mu_2^{(i)})\) and symmetric covariance matrix \[\Sigma_i = \left(\begin{array}{cc}\sigma_{1,1}^{(i)} & \sigma_{1,2}^{(i)} \\ \sigma_{1,2}^{(i)} & \sigma_{2,2}^{(i)}\end{array}\right).\] The known classes are 0-3.
library(mvtnorm)
set.seed(1)
# sample sizes
n_labeled <- 400
n_unlabeled <- 5000
n_feats <- 2
# mean values and covariance matrices
mu <- t(replicate(
10,
runif(2, -10, 10)
))
sigma <- t(replicate(
10,
{
A = matrix(runif(2^2)*2-1, ncol=2)
return(as.vector( t(A) %*% A))
}
))
rownames(mu) <- rownames(sigma) <- 0:9
print(mu)
#> [,1] [,2]
#> 0 -4.689827 -2.55752201
#> 1 1.457067 8.16415580
#> 2 -5.966361 7.96779370
#> 3 8.893505 3.21595585
#> 4 2.582281 -8.76427459
#> 5 -5.880509 -6.46886495
#> 6 3.740457 -2.31792564
#> 7 5.396828 -0.04601516
#> 8 4.352370 9.83812190
#> 9 -2.399296 5.54890443
print(sigma)
#> [,1] [,2] [,3] [,4]
#> 0 1.0873223 0.69488058 0.69488058 0.6528557
#> 1 0.2686249 0.50666811 0.50666811 1.0024862
#> 2 0.6486391 -0.09008240 -0.09008240 0.0409379
#> 3 0.3940044 -0.21990520 -0.21990520 0.5422173
#> 4 0.9611412 0.40244041 0.40244041 0.2316753
#> 5 0.4985329 0.39442301 0.39442301 0.3314552
#> 6 0.3384411 -0.08302008 -0.08302008 0.9109265
#> 7 0.3644605 0.25766878 0.25766878 0.5238927
#> 8 0.2758416 0.51517414 0.51517414 1.3789753
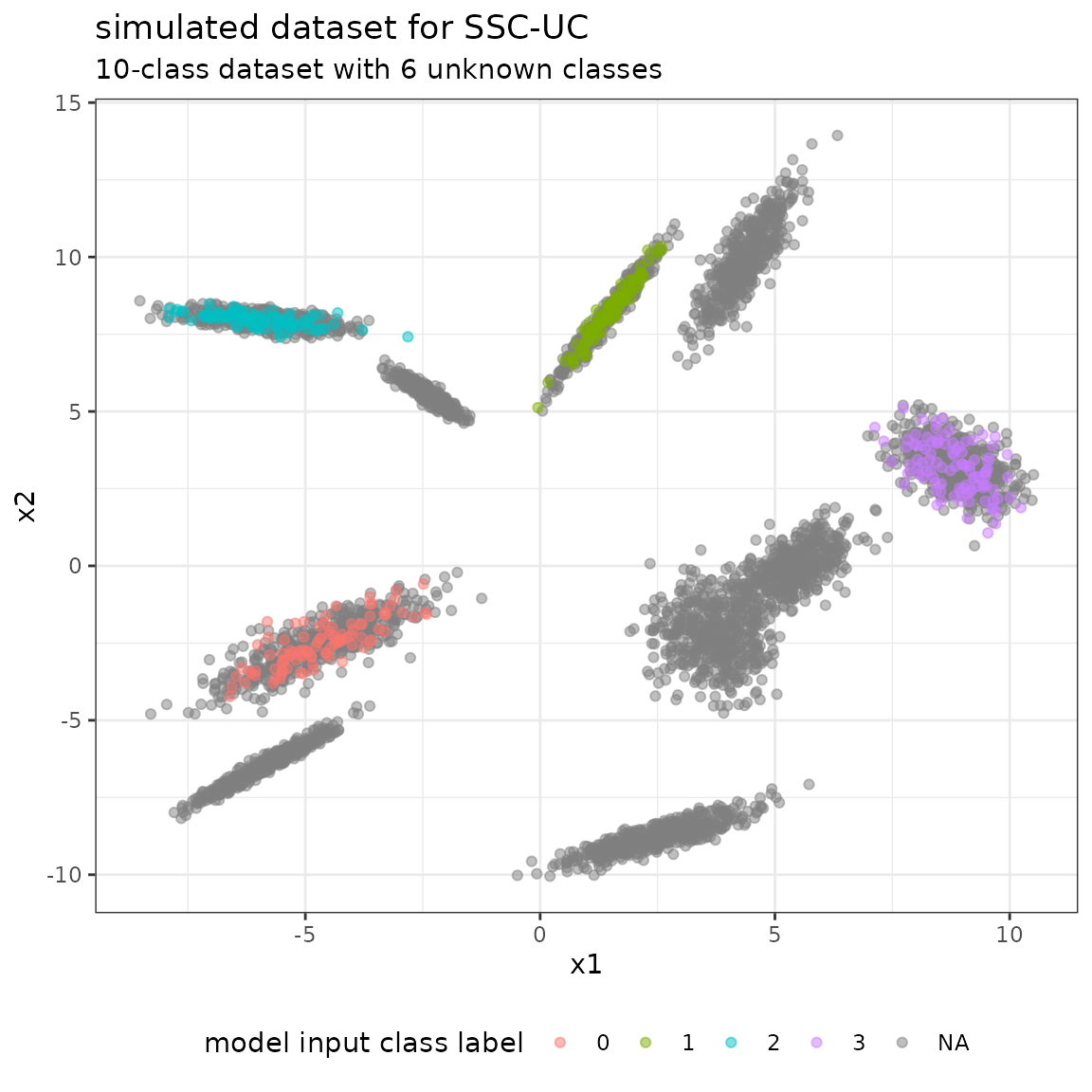
#> 9 0.1364133 -0.12600427 -0.12600427 0.1397050Given the data specifications, we simulate from the bivariate Gaussian distribution to generate the dataset given the class vector. We discriminate between the true class vector y_true containing the labels 0-9 of all samples, and the model input class vector y containing a true label only for the labeled data, and NA, otherwise.
# specify number of labeles / unlabeled samples per class
labeled_classes <- rep(0:3, each = n_labeled / 4)
unlabeled_classes <- rep(0:9, each = n_unlabeled / 10)
num_sample <- c(table(labeled_classes), table(unlabeled_classes))
# simulate X, ytrue and y
X <- c()
for(i in 1:length(num_sample)){
X <- rbind(X, rmvnorm(num_sample[i],
mu[names(num_sample)[i],],
matrix(sigma[names(num_sample)[i],], nrow = 2, ncol = 2)
)
)
}
y <- rep(c(0:3, NA), c(table(labeled_classes), length(unlabeled_classes)))
ytrue <- rep(names(num_sample), num_sample)
colnames(X) <- paste0("x", 1:2)
# summaries of the model input class vector y, and the true class vector ytrue
summary(as.factor(y))
#> 0 1 2 3 NA's
#> 100 100 100 100 5000
summary(as.factor(ytrue))
#> 0 1 2 3 4 5 6 7 8 9
#> 600 600 600 600 500 500 500 500 500 500Using the model input class vector y and the simulated feature matrix X, we generate the input dataset and the formula for the model.
# input dataset for the model
data <- as.data.frame(cbind(X, y))
# model formula
formula <- y ~ x1 + x2 - 1
# simulated data
head(data)
#> x1 x2 y
#> 1 -3.438659 -2.052945 0
#> 2 -4.343196 -2.430395 0
#> 3 -6.177755 -3.426197 0
#> 4 -5.090663 -2.765295 0
#> 5 -3.318454 -1.566550 0
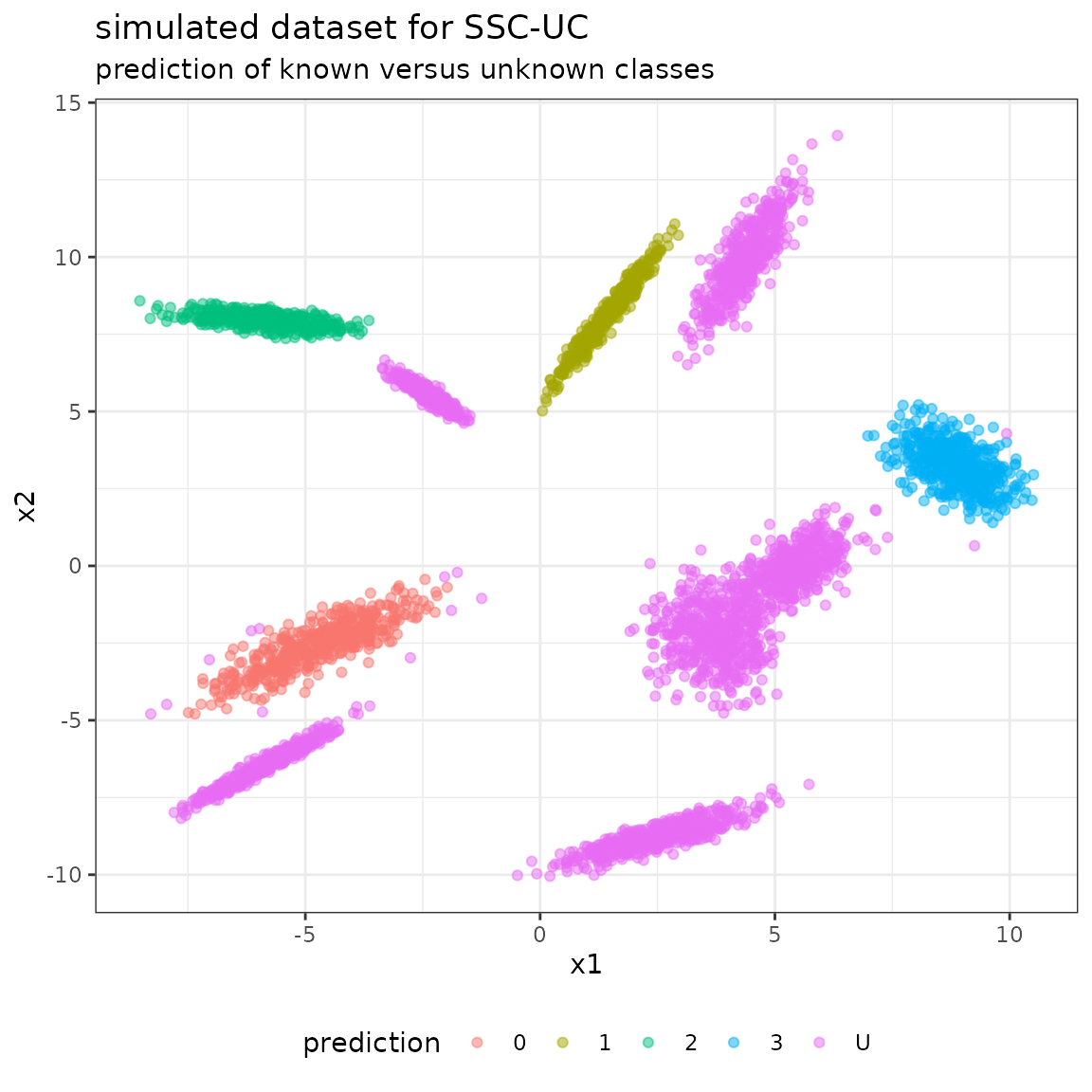
#> 6 -4.953884 -2.801539 0The simulated dataset is given by the following scatterplot:
Train SSC-UC model
The SSC-UC model is called via SSCUC and returns a BayesClassifier object with known and unknown classes. In this case, a Bayes classifier is used, specified by the argument naive=TRUE.
library(SSCUC)
# train model
model <- SSC(formula, data, naive = FALSE)
#> Warning in summary.mclustBIC(BIC, data, G = G, modelNames = modelNames): best
#> model occurs at the min or max of number of components considered!
#> Warning in Mclust(X[newclass_inds, !const_cols, drop = F], G = g_opts,
#> modelNames = gmmModelName, : optimal number of clusters occurs at max choice
#> [1] "Starting EM with 6 classes"
#> Warning in BayesClassifier(formula, data, naive = naive, prior = prior, :
#> BayesClassifier removed 1957 NAs
#> [1] "EM converged after 5 iterations"
#> [1] "BIC: 18418.8851091367"
#> [1] "Trying EM with 5 classes"
#> Warning in BayesClassifier(formula, data, naive = naive, prior = prior, :
#> BayesClassifier removed 1957 NAs
#> [1] "EM converged after 4 iterations"
#> [1] "BIC: 22778.8389840301"
#> [1] "BIC increased when updating - stopping"
summary(model)
#> BayesClassifier model with 10 classes and 2 non-constant features
#> ==============================
#> formula: y ~ x1 + x2 - 1
#> used features: x1, x2
#> parameters:| mu | Sigma | prior | |
|---|---|---|---|
| 0 | -4.66,-2.53 | 0.84,0.52,0.52,0.5 | 0.09 |
| 1 | 1.47,8.19 | 0.27,0.48,0.48,0.96 | 0.11 |
| 2 | -5.94,7.96 | 0.67,-0.09,-0.09,0.05 | 0.11 |
| 3 | 8.81,3.25 | 0.31,-0.18,-0.18,0.44 | 0.09 |
| U1 | 2.51,-8.77 | 0.96,0.4,0.4,0.24 | 0.09 |
| U2 | 4.55,-1.18 | 1.08,1.06,1.06,1.93 | 0.18 |
| U3 | -5.88,-6.47 | 0.56,0.45,0.45,0.4 | 0.09 |
| U4 | -2.4,5.55 | 0.13,-0.11,-0.11,0.13 | 0.09 |
| U5 | 4.37,9.92 | 0.27,0.5,0.5,1.37 | 0.09 |
| U6 | 0.29,1.24 | 43.47,11.45,11.45,17.91 | 0.05 |
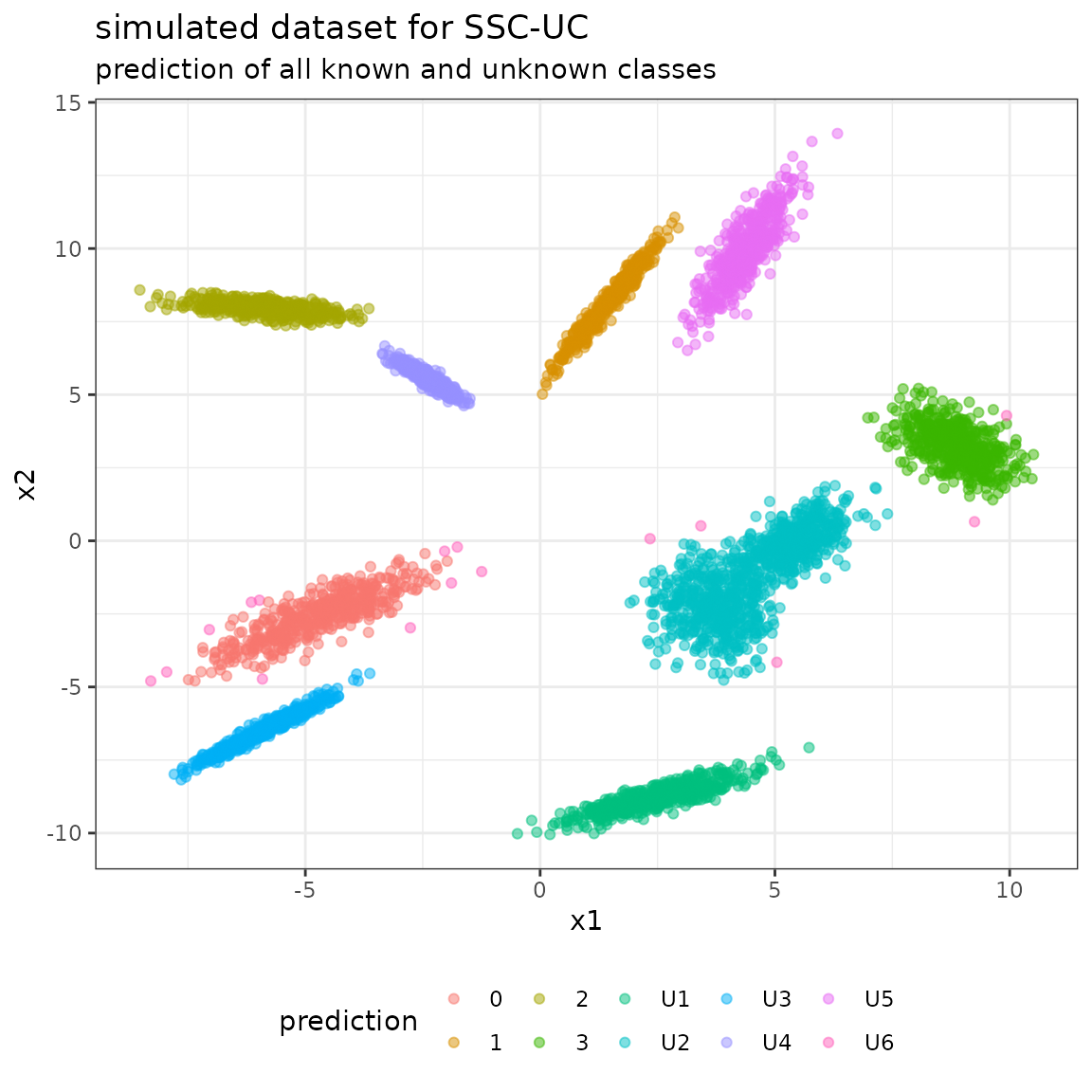
Predict and evaluate unlabeled data using SSC-UC model
Class labels for the unlabeled data in the dataset are obtained using the predict function with argument type=“class”.
# predict on unlabeled data
pred <- predict(model, newdata = subset(data, is.na(y)), type = "class")Evaluate in full confusion matrix (multiple unknown classes)
The full confusion matrix contains all the classes modeled by the BayesClassifier:
- the class labels 0-9 from the data specification as reference labels (not that, however, only classes 0-3 are known to the model a-priori),
- the class labels for known classes (0-3) and unknown classes (U1-U8) as predicted labels.
library(caret)
#> Loading required package: lattice
library(knitr)
# specify levels
levels <- sort(union(unique(pred), unique(ytrue)))
kable(confusionMatrix(
reference = factor(ytrue[is.na(y)], levels = levels),
data = factor(pred, levels = levels))$table[sort(unique(pred)),sort(unique(ytrue))]
)| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 489 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 500 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 500 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 498 | 0 | 0 | 0 | 0 | 0 | 0 |
| U1 | 0 | 0 | 0 | 0 | 500 | 0 | 0 | 0 | 0 | 0 |
| U2 | 0 | 0 | 0 | 0 | 0 | 0 | 497 | 500 | 0 | 0 |
| U3 | 0 | 0 | 0 | 0 | 0 | 500 | 0 | 0 | 0 | 0 |
| U4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 500 |
| U5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 500 | 0 |
| U6 | 11 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 0 |
The following plot shows the unlabeled data with their predicted class labels:
Evaluate in reduced confusion matrix (all unknown classes as class “U”)
As a second evaluation step, we can summarize all “unknown” classes detected by SSC-UC into one class “U” (unknown). Thereby, we reduce the number of labels in the confusion matrix to 0-3 and U.
# set all new class labels to "U"
pred[!pred %in% unique(ytrue)] <- "U"
# specify levels
levels <- sort(union(unique(pred), unique(ytrue)))
kable(confusionMatrix(
reference = factor(ytrue[is.na(y)], levels = levels),
data = factor(pred, levels = levels))$table[sort(unique(pred)),sort(unique(ytrue))]
)| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 489 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 500 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 500 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 498 | 0 | 0 | 0 | 0 | 0 | 0 |
| U | 11 | 0 | 0 | 2 | 500 | 500 | 500 | 500 | 500 | 500 |
The following plot shows the unlabeled data with their predicted class labels, when considering all unknown classes as one class.